Comparing the Wilcoxon-Mann-Whitney to alternative statistical tests
2021-08-24
bioqc-wmw-test-performance.RmdIn this document, we show that the Wilcoxon-Mann-Whitney test is comparable or superior to alternative methods.
Two alternative methods could be compared with the Wilcoxon-Mann-Whitney (WMW) test proposed by BioQC: the Kolmogorov-Smirnov (KS) test, and the Student’s t-test, or more particularly, the Welch’s test which does not assume equal sample number or equal variance, which is appropriate in the setting of gene expression studies.
- It is documented in statistics literature that the WMW test offers a higher power than the Kolmogorov-Smirnov test1,2.
- Compared with parameterized test methods such as the t-test, the WMW test is (a) resistance to monotone transformation, (b) suffers less from outliers, and (c) provides higher efficiency when many genes are profiled and the distribution of gene expression deviates from the normal distribution, which are important criteria in genome-wide expression data.
Based on these considerations, BioQC implements a computationally efficient version of the WMW test. In order not to confuse end-users, no alternative methods are implemented.
Nevertheless, in order to demonstrate the power of WMW test in comparison with the KS-test or the t-test, we performed the sensitivity benchmark described in the simulation studies, for the two alternative tests respectively.
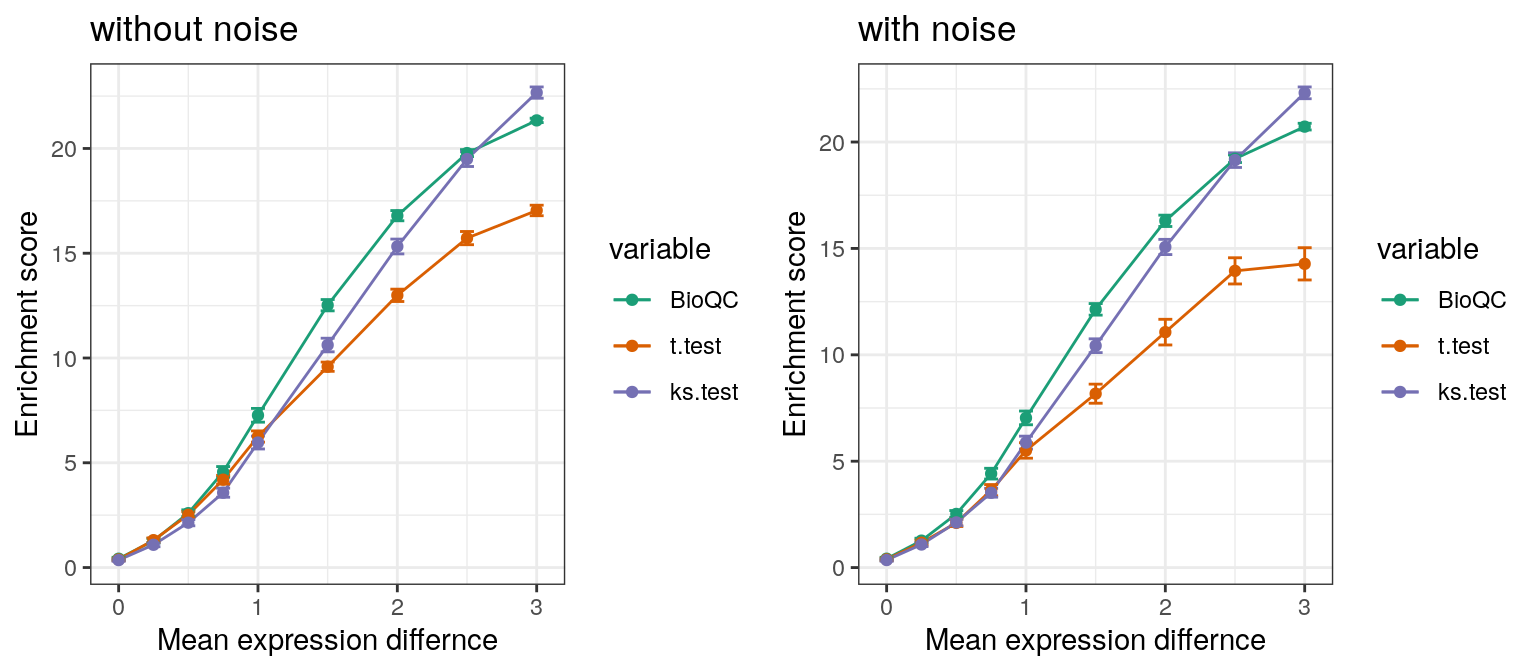
Figure 1: Sensitivity benchmark. Expression levels of genes in the ovary signature are dedicately sampled randomly from normal distributions with different mean values. The lines show the enrichment score for the Wilcoxon-Mann-Whitney test, the t-test and the Kolmogorov-Smirnov test respectively. In the right panel, outliers were added by adding a random value to 1% of the simulated genes.
As expected, the results suggest, that both the KS-test and the WMW-test are robust to noise, while the performance of the t-test drops significantly on noisy data. Additionally, the WMW-test appears to be superior to the KS-test for low expression differences.
Computational Performance
Since the KS-test is so slow, we did not replicate the sensitivity benchmark from the simulation studies using the enrichment score rank. While it takes BioQC about 4 seconds on a single thread to test all 155 signatures, it already takes the KS-test about 2 seconds to test a single signature.
## test replications elapsed relative
## 2 runKS() 5 10.528 1.00
## 1 runWMW() 5 18.635 1.77R Session Info
## R version 4.1.0 (2021-05-18)
## Platform: x86_64-pc-linux-gnu (64-bit)
## Running under: Ubuntu 20.04.2 LTS
##
## Matrix products: default
## BLAS/LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.8.so
##
## locale:
## [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
## [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
## [5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=C
## [7] LC_PAPER=en_US.UTF-8 LC_NAME=C
## [9] LC_ADDRESS=C LC_TELEPHONE=C
## [11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
##
## attached base packages:
## [1] stats4 stats graphics grDevices utils datasets methods
## [8] base
##
## other attached packages:
## [1] rbenchmark_1.0.0 ggplot2_3.3.5 plyr_1.8.6
## [4] reshape2_1.4.4 gplots_3.1.1 gridExtra_2.3
## [7] latticeExtra_0.6-29 lattice_0.20-44 hgu133plus2.db_3.13.0
## [10] org.Hs.eg.db_3.13.0 AnnotationDbi_1.55.1 IRanges_2.27.2
## [13] S4Vectors_0.31.1 BioQC_1.21.4 Biobase_2.53.0
## [16] BiocGenerics_0.39.2 testthat_3.0.4 knitr_1.33
##
## loaded via a namespace (and not attached):
## [1] bitops_1.0-7 fs_1.5.0 bit64_4.0.5
## [4] RColorBrewer_1.1-2 httr_1.4.2 rprojroot_2.0.2
## [7] GenomeInfoDb_1.29.3 tools_4.1.0 bslib_0.2.5.1
## [10] utf8_1.2.2 R6_2.5.0 KernSmooth_2.23-20
## [13] DBI_1.1.1 colorspace_2.0-2 withr_2.4.2
## [16] tidyselect_1.1.1 bit_4.0.4 compiler_4.1.0
## [19] textshaping_0.3.5 desc_1.3.0 labeling_0.4.2
## [22] sass_0.4.0 caTools_1.18.2 scales_1.1.1
## [25] pkgdown_1.6.1.9001 systemfonts_1.0.2 stringr_1.4.0
## [28] digest_0.6.27 rmarkdown_2.10 XVector_0.33.0
## [31] jpeg_0.1-9 pkgconfig_2.0.3 htmltools_0.5.1.1
## [34] highr_0.9 fastmap_1.1.0 limma_3.49.4
## [37] rlang_0.4.11 RSQLite_2.2.7 farver_2.1.0
## [40] jquerylib_0.1.4 generics_0.1.0 jsonlite_1.7.2
## [43] gtools_3.9.2 dplyr_1.0.7 RCurl_1.98-1.3
## [46] magrittr_2.0.1 GenomeInfoDbData_1.2.6 Rcpp_1.0.7
## [49] munsell_0.5.0 fansi_0.5.0 lifecycle_1.0.0
## [52] stringi_1.7.3 yaml_2.2.1 edgeR_3.35.0
## [55] zlibbioc_1.39.0 grid_4.1.0 blob_1.2.2
## [58] crayon_1.4.1 Biostrings_2.61.2 KEGGREST_1.33.0
## [61] locfit_1.5-9.4 pillar_1.6.2 glue_1.4.2
## [64] evaluate_0.14 png_0.1-7 vctrs_0.3.8
## [67] gtable_0.3.0 purrr_0.3.4 cachem_1.0.5
## [70] xfun_0.25 ragg_1.1.3 tibble_3.1.3
## [73] memoise_2.0.0 ellipsis_0.3.2