Discoveries Weekly No. 13 (July 6-July 12, 2020)
Here are some discoveries that fascinate me this week.
- Drug discovery
- Computational biology
- Programming
- Statistics
- Career paths of programmers: on the ladder, on the ground, and the third way
- Other gems
Drug discovery
ADME properties of antibodies
I found an interesting review about adsorption, distribution, metabolism, and excretion (ADME) properties of biologics, focusing especially on the learnings from small molecules: Thomayant Prueksaritanont and Cuyue Tang, AAPS Journal, 2012), thanks to recommendation by colleagues. A related paper, Jain et al., PNAS, 2017, examines biophysical properties of the clinical-stage antibodies, which I would like to imagine as ‘survivors’ of discovery and development programs, and provides a complementary view on this topic.
On the antibody side, many antibodies follow the structure of Immunoglobulin G (IgG) antibodies, the most common type of antibody found in circulation and extracellular fluids. IgG consists of both heavy and light chains. Besides IgG, there are other four heavy-chain isotypes, known as IgA, IgD, IgE, and IgM. With regard to light chain, there are two isotypes: $\kappa$ and $\lambda$. Here, isotype (class) means the unique constant region segments of the immunoglobulin gene which form the fragment crystallizable (Fc) region and the lower segment of the fragment antigen-binding (Fab) portion of an antibody. More about isotypes can be found on Wikipedia.
There are four subclasses of IgGs (IgG1-4). IgG1 (crossing placenta, complement activation, and high affinity to Fc receptor on phagocytic cells), IgG2 (not crossing placenta, moderate complement activation, but extremely low affinity), and IgG4 (crossing placenta, no complement activation, and intermediate affinity) are often chosen as the antibody formats. Many variants and formats of antibodies are available nowadays, which are reviewed by Spiess et al..
IgG alone can protect the body from infection through the activities of its antigen binding region. However, immune functions of IgGs are much mediated by proteins and receptors that binding to the Fc region of IgG, known collectively as Fc receptors. Fc receptors can be classified into three classes by the antibody type that they bind to: Fc-gamma receptors (binding to IgG antibodies), Fc-alpha receptors (binding to IgA antibodies), and Fc-epsilon receptors (binding to IgE antibodies).
Fc-gamma receptors (Fc$\gamma$R) receptors contain many classical membrane-bound surface receptors, as well as atypical intracellular receptors and cytoplasmic glycoproteins. A particular atypical Fc$\gamma$R, the neonatal Fc receptor (FcRn), is particularly interesting, among others because it can strongly influence IgG biology, including stability and PK/PD profiles of IgG format antibodies and albumin. It acts as a recycling or transcytosis receptor that maintains IgG in the circulation, and transport them across cellular barriers. It is also an immune receptor by interacting with and assisting antigen presentation of peptides derived from IgG immune complexes. Two interesting reviews about biology of FcRn can be found in Roopenian and Akilesh, Nature Reviews Immunology, 2007 and in Pyzik, et al., Front. Immunol., 2019.
Multispecific drugs
I summarized my learning notes in another post.
Computational biology
Applied bioinformatics for the identification of regulatory elements
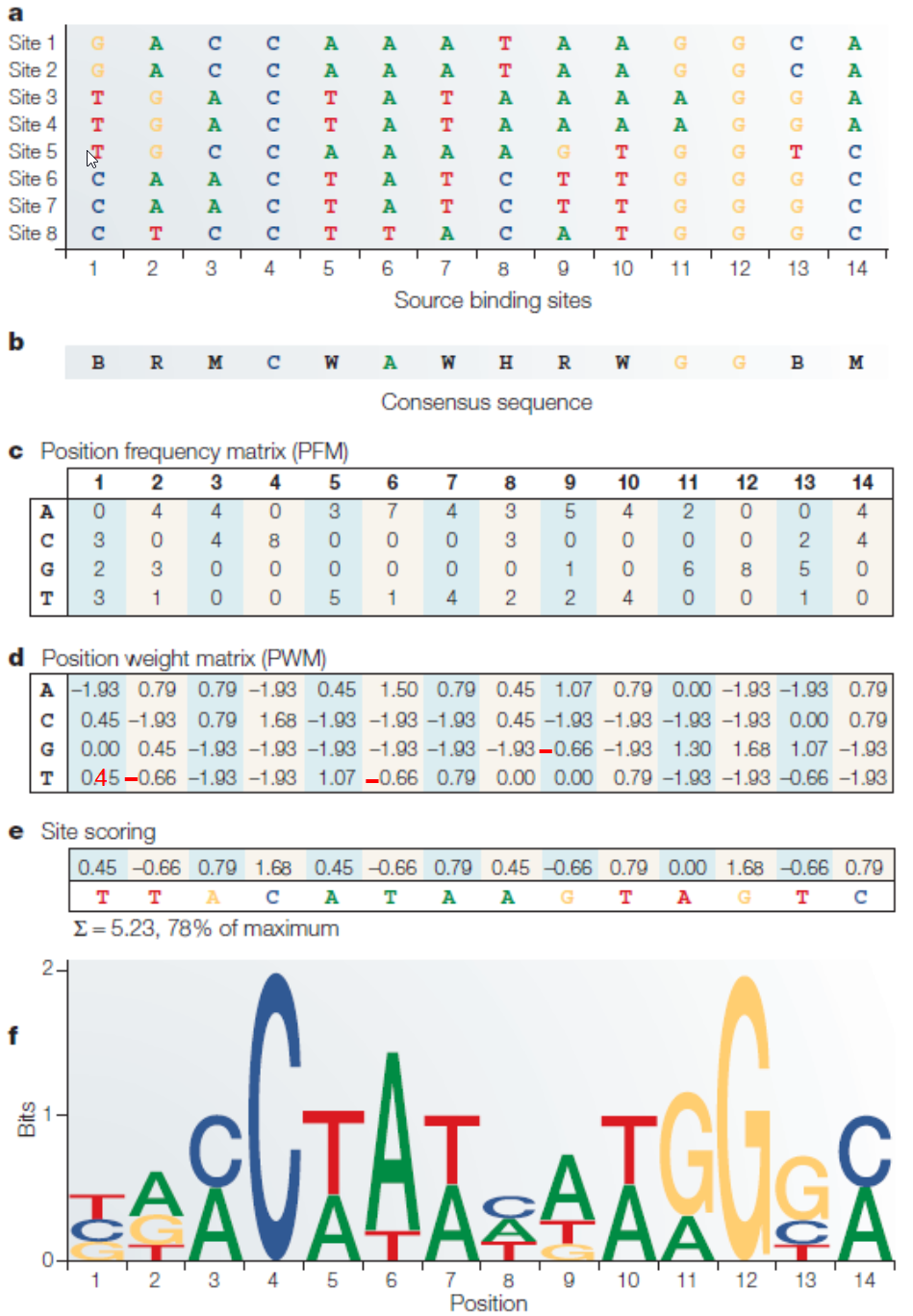
Wasserman and Sandelin, Nature Review Genetics, 2004 is a classic paper in the field of bioinformatics and genomics. It introduced key concepts such as position weight matrix (PWM), also known as position-specific score matrix (PSSM), phylogenetic footprinting (identification of conserved regulatory elements by comparing genomic sequences between related sequences), as well as combinatorial interaction of transcription factors via cis-regulatory module (CRM) analysis. These and other concepts introduced in the paper are fundamental for many tools that we use today to characterize elementary features in genomes.
In Box 1 of the paper, the authors gave an example on how to derive a PWM from a set of aligned sequences. Unfortunately, there seem to be a few typos in the tables, which obscures the interpretation. Here are some that I found:
Programming
Simple graphical user interfaces with guietta and Python
alfiopuglisi/guietta is a tool making simple graphical user interfaces (GUIs) with Python. Discovered from hacker news.
Statistics
Hill tail-index estimator and Hill equation
The Hill tail-index estimator was proposed by Bruce M. Hill as a simple and general method to inference about the tail behaviour of a distribution (Hill et al.). It does not assume any global form for the distribution function, but only the form of the behaviour in the tail where it is desired to draw inference. It is a way to describe heavy-tailed distributions, besides Pickand’s and Ratio estimators of the tail-index.
Do not mix it with the Hill coefficient in Hill equation in biochemistry, which is attributed to the English physiologist Archibald Vivian (A.V.) Hill. As the Hill coefficient increases, the saturation curve becomes steeper.
Career paths of programmers: on the ladder, on the ground, and the third way
About the news Linux Tovarlds: I am not a programmer anymore, here are a few interesting views on Slashdot:
From guruevi, an apparently good manager:
It’s the (good) evolution of any technical manager - you’ve got too much work to delve deep into the code and its dependencies. I haven’t learned much new programming languages either in the last few years, I just farm the work out to my minions, write in pseudocode and once in a while I will read the documentation of some new framework or library.
The rest of my time is spent dealing with superiors and customers, mostly managing expectations and shielding my team from the ire of some micromanager.
From jd, a programmer,
I consider programming enjoyable, not a chore. Couldn’t give a damn if I’m still coding into my 90s. I like solving problems and that’s a category of problem.
Spend my time away from keyboards solving different problems for other organizations. Archaeology, history, maths, I don’t care, it’s call problem solving and all fun.
From TechyImmigrant, a hardware architect with programming skills
In my job, people regard me as a hardware architect more than coder (of System Verilog RTL, python and C mostly).
This comes from years of coding, during which I developed some important circuits for my employer with cunning designs.
Then they promote you and want you to write documents describing things to be coded by others.
I find that problematic, because all my most cunning designes were arrived at iteratively, coding up solutions, identifying problems and then refining the solution until it worked for being coded, its size and efficiency, debugability, testability on the lab bench and in high volume manufacturing and solving the problems of remaining secure while remaining testable.
So I still code RTL and Python and C when coming up with my designs, document them and them throw the code and documents over to the rest of the team to beat it into submission, test it and help kick it into shape for mass production.
Apparently these are the three typical career paths of programmers: becoming professional managers (which I call ‘on the ladder’), continue coding and solving problem (‘on the ground’), and trying to stay between the two (with one foot on the ladder and the other on the ground).
I can imagine that these career paths are also the ones scientists and engineers can choose when they work in industry. Each and everyone of us is asked to make a decision based on circumstances and his capability and wishes.
Other gems
- I read this week the book Man’s Search for Meaning by Victor Frankl. The conclusion I draw is that we do not ask life for a meaning, rather life asks us for a meaning. Given circumstances, we have to make active choices and live a life that we think worth living.
- Math book recommendations for teenagers, discussion on hacker news.
- Design thinking for data products by Norbert Wirth and Martin Szugat.
- Wir haben das Hier und Jetzt. Und das ist alles, was wirklich zählt (We have Here and Now. This is all that really matters), an interview with Prof. Dr. Andrew Gloster (in German).
Happy weekend!