This is an archived page of the AMIDD course 2023. Please visit the course’s website for the current version.
Welcome to the website for Applied Mathematics and Informatics In Drug Discovery (AMIDD), the course series running at the Department of Mathematics and Informatics, University of Basel in the fall semester 2023.
The course series introduces interdisciplinary research in drug discovery with mathematics as the language and computation as the tool. We have a diverse and lively class room that learn together and from each other: every year about two third students of the class study mathematics or computer science, while other students study physics, chemistry, (computational) biology, pharmacy, and other fields such as epidemiology and medicine.
More information on the course can be found at the course directory of the University Basel.
Table of content
- Time and place
- Course material and licensing
- Pre-course survey
- Assessment
- Syllabus
- Lecture 1: Introduction (22.09.2023)
- Lecture 2: Protein as drug targets and mathematical modelling (29.09.2023)
- Lecture 3: Statistical modelling and machine learning (06.10.2023)
- Lecture 4: Causal inference (13.10.2023)
- Lecture 5: Searching for key questions for drug discovery (20.10.2023)
- Lecture 6: Five key questions of drug discovery (27.10.2023)
- Lecture 7: Biological sequence analysis (03.11.2023)
- Lecture 8: Ligand-based and structure-based drug discovery (10.11.2023)
- Lecture 9: From interactions to networks (Attention Room Change) (17.11.2023)
- Dies academicus (24.11.2023, no lecture)
- Lecture 10: Omics and MoA studies for translational research (01.12.2023)
- Lectuer 11: PK/PD modelling (08.12.2023)
- Lecture 12: Guest lectures (15.12.2023)
- Lecture 13: A collaboration challenge (22.12.2023)
- Further questions or suggestions?
- Offline activities
- Archives of past courses
Time and place
The lecture takes place on Fridays between 12:15 and 14:00 at Spiegelgasse 5, Seminarraum 05.002.
Course material and licensing
Course material, including lecture notes, slides, and reading material, are shared on the course’s web site, AMIDD.ch, unless otherwise specified in the course.
All course material, unless otherwise stated, is shared under the Creative Commons (CC-BY-SA 4.0) license.
Pre-course survey
Prior to attending the first session, please fill out the voluntary pre-course survey. Your reply helps me to shape the course to meet your needs.
Assessment
The final note is given by participation including quizzes (30%), offline activities (40%), and a collaboration challenge in the final session (30%).
Syllabus
Lecture 1: Introduction
The first module is finished with the first lecture.
- Slides
- Please fill the anonymous post-lecture survey of Lecture 1.
- Offline activities of Lecture 1: Check out this poster, which illustrates top 200 brand name drugs by retail sales in 2022. You can also find a snapshot in the slide deck. Focus on the top 14 compounds, and answer the questions by filling out this form.
Lecture 2: Drug targets and mechanistic modelling
- Slides of lecture 2 on drug targets and mechanistic modelling.
- Please fill the anonymous post-lecture survey of Lecture 2.
- Offline activities of Lecture 2:
- (Optional) Watch the video tutorials hyperlinked in the slides if you want to get familiar with protein structure and protein function classes.
- Watch a video on the development of the drug Herceptin, presented by Susan Desmond-Hellmann and answer questions. See the questions and submit your answers here via a Google Form.
- Required reading: Principles of early drug discovery by Hughes et al.
Lecture 3: Statistical modelling and machine learning
- Slides of lecture 3 on statistical modelling and machine learning.
- Offline activities of Lecture 3: read the following papers and answer the
questions via Google Form. The deadline
of submitting your answers is by the end of lecture 5.
- Required reading 1: An introduction to machine learning by Badillo et al.
- Required reading 2: The Environment and Disease: Association or Causation? by Bradford Hill.
Lecture 4: Causal inference
- Slides of lecture 4 on causal inference.
- Offline activities: read the review on causal inference and its application in drug discovery and development. Answer the questions via Google Form. The deadline of submission is by the end of lecture 6.
Lecture 5: Searching for key questions in drug discovery
- Slides of lecture 5. In this lecture, we played roles to search for key questions for drug discovery.
- Offline activities: read Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma by Bollag et al., Nature 2010. Answer questions by filling out this Google Form. The activities must be finished before lecture 7.
Lecture 6: Disease understanding and target identification
- Slides of lecture 6.
- Offline activities: continue reading the paper assigned last week, and fill the Google Form.
Lecture 7: Biological sequence analysis
- Slides of lecture 7. We finished the content until WebLogo. Markov chain, hidden Markov chain, and convolutional neural network will be discussed in the next lecture.
- Offline activities (no need to report via Google Form, except for the feedback)
- Fill out a feedback form to this lecture and for the lectures so far.
- Check out the examples of dynamic programming and constant-space solution of the Fibonacci problem, and make sure that you can implement them yourself.
- Calculate the Levenshtein distance between
ATTAATGCCandATATTTCGCCusing dynamic programming, and validate your results with a programming language or large-language model of choice.
Lecture 8: Structure-based and ligand-based drug discovery
- Slides of lecture 8.
- Offline activities: Submit the results before November 24th to the Google form.
- Compare
p(ACGTGGT|M)andp(ACCTGGT|M), whereMstands for the Markov model given in the slide, in the Google form, and at the end of this section. - We have got a RNA sequence by sequencing sputum from a patient (see below). How can we know the original genome of the sequence, and ideally the gene encoding the sequences? Tips: go to the NCBI BLAST tool, copy and paste the sequence as the query sequence, and try your luck. Default parameters are okay. The query sequence is:
ATGTTTGTTTTTCTTGTTTTATTGCCACTAGTCTCTAGTCAGTGTGTTAATCTTACAACCAGAACTCAATTACCCCCTGCATACACTAATTCTTTCACACGTGGTGTTTATTACCCTGACAAAGTTTTCAGATCCTCAGT. - Required reading: Selected pages from Evaluation of the Biological Activity of Compounds: Techniques and Mechanism of Action Studies by Iain G. Dougall and John Unitt, chapter two of the book The Practice of Medicinal Chemistry. To answer offline-activity questions, it is required to read pages 15-22 (1-8 of the 29 pages in total, before section ‘4. Types of Enzyme Inhibition and Their Analysis’), page 27 (section 6A), and pages 34-37 (Assay Biostatistics). The rest is optional reading.
- Compare
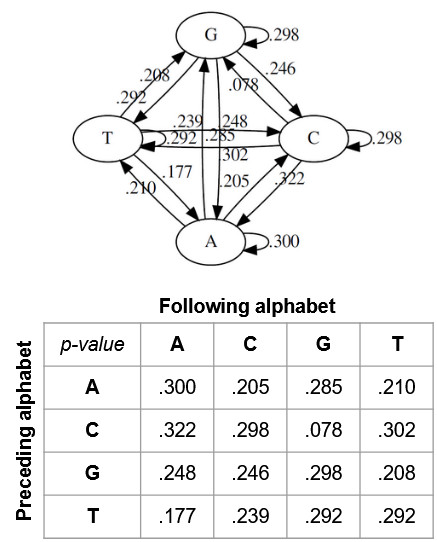 .
.
Lecture 9: From interactions to networks
The course takes place exceptionally at Bernoullistrasse 30/32, kleiner Hörsaal 120.
- Slides of lecture 9.
- Offline activities: read Computational methods in drug discovery by Gregory Sliwoski et al. Please submit your replies to questions via this Google Form. The deadline of submitting the replies is December 1st.
Note that on Friday, 24.11.2023, there is NO lecture due to Dies Academicus.
Lecture 10: Omics and MoA studies for translational research
- Slides of lecture 10.
- Offline activities: read Ten Quick Tips for Effective Dimensionality Reduction by Nguyen und Holmes. Apply the knowledge to a real-world example: See the instructions in the slides.
Lecture 11: PK/PD modelling
- Slides of lecture 11.
- Required reading: pick one publication to read according to your interest. No reply submission required.
- [Industrial approach to predict human PK] Davies, Michael, et al.. 2020. “Improving the Accuracy of Predicted Human Pharmacokinetics: Lessons Learned from the AstraZeneca Drug Pipeline Over Two Decades.” Trends in Pharmacological Sciences 41 (6): 390–408.
- [Introduction to PBPK modelling] Jones, H. M., and K. Rowland‐Yeo. 2013. “Basic Concepts in Physiologically Based Pharmacokinetic Modeling in Drug Discovery and Development.” CPT: Pharmacometrics & Systems Pharmacology 2 (8): 63.
- [Application of machine learning for PK prediction] Stoyanova, R. et al. Computational Predictions of Nonclinical Pharmacokinetics at the Drug Design Stage. J. Chem. Inf. Model. 63, 442–458 (2023).
Lecture 12: Guest lectures
We shall have two guest speakers: Hanna Silber Baumann and Ercan Suekuer.
- Hanna Silber Baumann holds a PhD in pharmacometric modeling and simulation. She has been working at Roche in the modeling and simulation group for about 10 years. Her talk will be between 12:15 and 12:40.
- Ercan Suekuer holds a B.Sc. in Bioinformatics, a M.Sc. in Bioprocess Engineering, and a M.Sc. in Data Science (AI). He has spent the majority of his career since 2008 in the pharmaceutical industry. Now he is a Senior Data Scientist at Roche, where he provides data support to colleagues in both the preclinical and clinical environments. His talk will be between 12:45 and 13:10.
After the two talks, we will use the rest of the time to
- Review offline activities so far;
- Give instructions about the collaboration challenge of the last lecture;
- Fill the evaluation form of the course.
Lecture 13: A collaboration challenge
Details are announced in the classroom during lecture 12.
Offline activities
You can find all offline activity questions and answers for AMIDD 2023, as well as answers to the questions raised by participants, in this PDF document.
Further questions or suggestions?
Please contact the lecturer, Jitao David Zhang, at jitao-david.zhang@unibas.ch.